New PhageClouds database makes genomic comparisons efficient
New data resource enables more efficient placement of novel phage genomic sequences, drawing together data from a variety of databases and public virome assemblies. This enables easier exploration of phage diversity, potentially allowing for better use of phages as a therapeutic approach in the future.
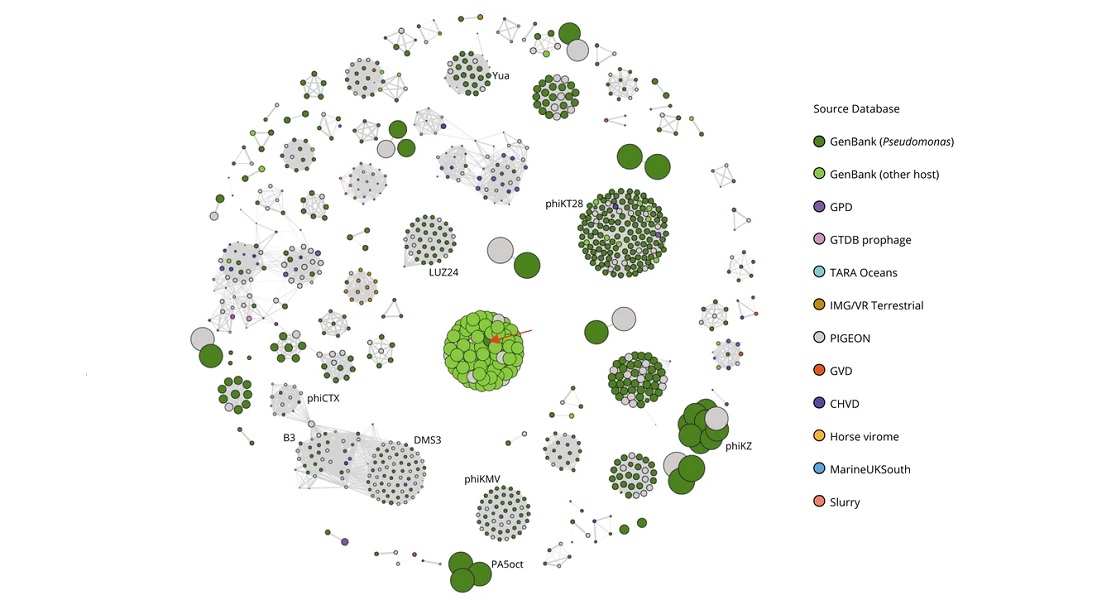
In line with the recent proliferation of research into bacteriophages and their therapeutic, biogeochemical, ecological, evolutionary and health potential, genomic data of these bacteriophages has exploded. So much so that the typical handling of data, alignment-based methods, have become impractical, as it now requires significant computing memory to handle the amount of data, thus making the genomic estimations unnecessarily slow. The new PhageClouds database attempts to solve this problem.
“PhageClouds is the GLOBE Institute’s digital phage collection and one of the largest world wide,” says Professor Thomas Sicheritz-Pontén from the Center for Evolutionary Hologenomics and continues: “It allows us to compare, categorise and select bacteriophages for making informed and rapid decisions about which phages we should combine in so-called Phage Cocktails to efficiently treat bacterial infections and invasions.”
PhageClouds has been set up by Professor Thomas Sicheritz-Pontén and Associate Professor Bent Petersen from the Center for Evolutionary Hologenomics alongside colleagues from the GLOBE Institute, including Guillermo Rangel-Pineros, postdoc at the Palaeoproteomics Group. Guillermo has recently first-authored an article about the advantages of PhageClouds in PHAGE: Therapy, Applications, and Research.
“In recent years the use of metagenomics in different environments has led to a very sharp expansion in the rate at which novel phage genomes are being reported. PhageClouds offers an alignment-free alternative that allows users to rapidly and accurately find phage genomic sequences closely related to their own phage genomes in a matter of seconds,” says Guillermo Rangel-Pineros.
Massive datasets for comparisons
PhageClouds contains the data from a total of 640,000 phage genomic sequences. This allows for researchers to explore phage diversity and compare between them. Especially important is the ability to search from a host-centric perspective, meaning that it allows for researchers to identify clusters of closely related phages that target a specific bacterial host, giving a visual overview of phages that are associated with it.
“This lets researchers contextualise their phage or group of phages of interest, i.e. to see what their phages are most closely related to, to see if there are lots of other phages in similar groups, and to see if they are related to other known phages that infect other bacterial species. This would enable us to acquire the knowledge required to inform the development of phages for therapeutic use,” says Guillermo Rangel-Pineros.
PhageClouds therefore, by utilising technological advancements to handle massive amounts of data, helps to open up the field of phages for the future. This is important, as phages have the potential to alleviate some pressing future medical problems, such as working to treat bacterial infections, which in light of the increase of antibiotic resistant pathogens may otherwise become problematic.
Read the full article published in PHAGE: Therapy, Applications, and Research here.
Contact:
Professor Thomas Sicheritz-Pontén, thomassp@sund.ku.dk
Postdoc Guillermo Rangel-Pineros, guillermo.pineros@sund.ku.dk
