The Plant Hologenome Group aims to uncover what makes plants resilient
Researchers from the Center for Evolutionary Hologenomics are working towards preventing future declines in plant-based food production in the face of climate change. The new Plant Hologenome Group, led by Assistant Professor Christopher Barnes, hopes to pinpoint resilient plant genotypes through hologenomic studies, which can then be used to breed stronger crops for the future.

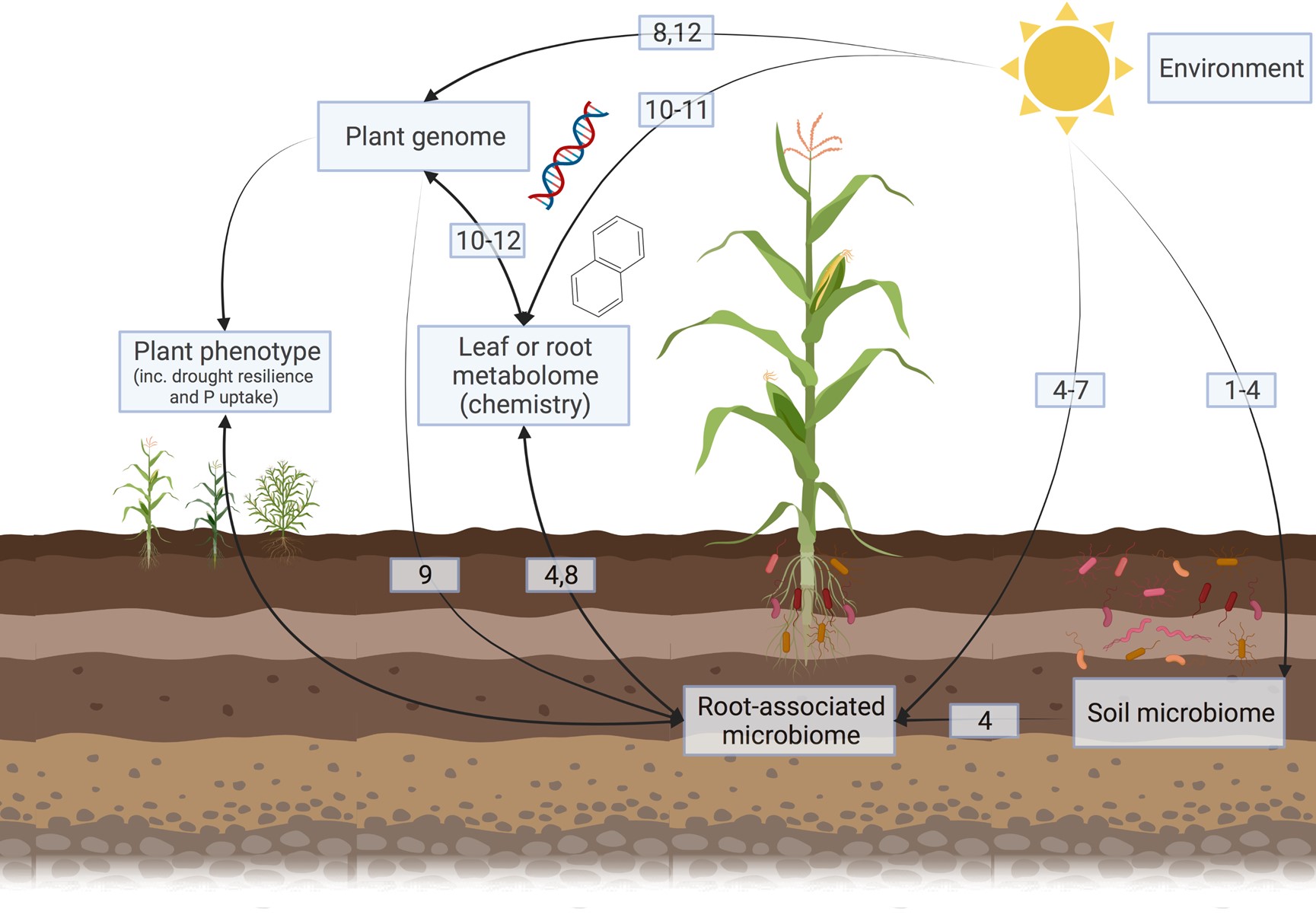
We’ve sat down with Assistant Professor Christopher James Barnes, who leads the new Plant Hologenome Group at the Center for Evolutionary Hologenomics (CEH), to talk about his work and aspirations for his new research group. The Plant Hologenome Group focuses on understanding phenotypic variations of plants using the holobiomic framework, meaning that the plants’ genome and microbiome together are considered to shape the functioning of the plant. The hope is that this research can improve drought-tolerance and pathogen resistance in crops.
“My work used to be typical of many other researchers, in that it was very fragmented, integrating only parts of the plant’s environment, genome and microbiome in attempting to understand the plant phenotype,” says Christopher Barnes and continues: “However, a few years ago, I came to the realization that these need to be studied simultaneously if we are to go beyond correlations to identify the causes and mechanisms behind variation in the plant phenotype. And that is why I established The Plant Hologenomics group, and that’s why I think the CEH is an awesome place to be.”
What can you discover by studying the plant phenotype?
“In an old project focusing on Plantago major (broadleaf plantain), I have linked genotypic variation to changing leaf chemistry and microbiome composition, demonstrating that the microbiome is influenced by the genotype, even across environmental gradients. This is why I believe that genotypic variation of the microbiome affects functioning, which can be exploited within crops (within breeding programmes or directly with genetic modification). Therefore, I am currently involved in larger applications to apply a multi’omic approach on barley and maize.”
Why is this important?
“While a better understanding of the plant phenotype sounds very theoretical, it has some very real implications. Climate change is increasing the frequency and intensity of weather extremes, such as drought. Drought tolerance is part of the plant phenotype, and I am particularly interested in studying the drought resilience of maize. Within Tanzania, for example, maize is the most important source of carbohydrates, but current models suggest yields could decline by up to 80% by 2050 without major interventions.”
How will you be moving forward?
“In my short-term, I am planning on applying for an ERC Starting grant to show that the root-associated microbiome of maize can be manipulated to affect the host’s drought tolerance (either positively or negatively). But my long-term aim is to determine whether some of the drought tolerance of the maize ancestor, teosintes, can be bred into modern maize to improve its drought resilience by manipulating the microbiome.”
How did you get started with this type of research?
“My background is as a microbial ecologist where I was interested in understanding the regulation of microbial communities, particularly associated with plants. As part of this, I performed many studies investigating the relationship between the environment (climate and soil properties) and the root-associated microbiome. More recently, I have performed studies finding genotypic variation of hosts (maize, broadleaf plantain) correlates to differences in the microbiome composition, even when grown in controlled environments. Similarly, I have been finding genotypic variation in the metabolome of these plants.”
