The Holo-omic approach opens up new research avenues
PhD student Lasse Nyholm Jessen and his colleagues from the HoloFood EU project and Center for Evolutionary Hologenomics at University of Copenhagen have published a perspective paper on the new possibilities of holo-omics in the open access journal iScience.
The new Holo-omic approach incorporates complex multi-omic data from both host and microbiota domains to untangle the interplay between the two. The authors highlight the wide range of holo-omic applications in varying research avenues such as biomedicine, biotechnology, agri- and aquacultural sciences, nature conservation as well as basic ecological and evolutionary research.
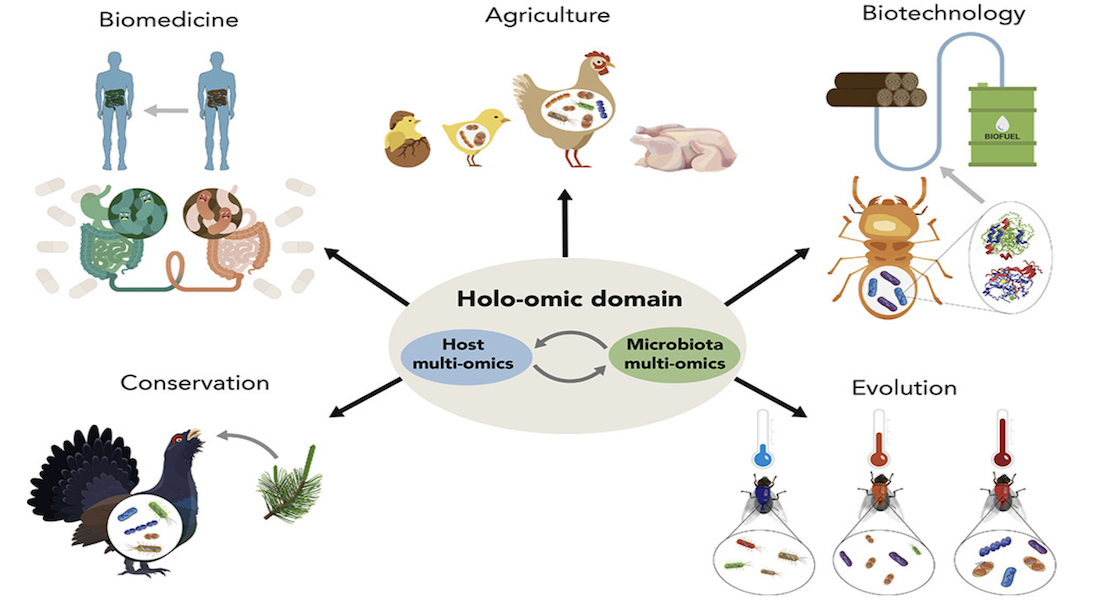
Holo-omics is essential to fully understand what shapes the phenotypes of complex organisms
Nyholm and colleagues point out the complexity and challenges of applying this new holo-omic approach. At the same time, the authors also highlight the importance of understanding these bi-directional interactions between the host organisms and their associated microbiota in order to understand what shapes the phenotypes of complex organisms. They do this by revisiting available evidence for the biological importance of host-microbiota interactions.
The relationship and effect of the environment on both host and respective microbiome and the interaction between the latter remains largely in the dark, especially in the context of wild organisms where the debate of what constitutes a healthy microbiome is still undecided.
The authors note: “This highlights the relevance of acknowledging and understanding the biomolecular interactions occurring between different omic levels of hosts and microorganisms.”
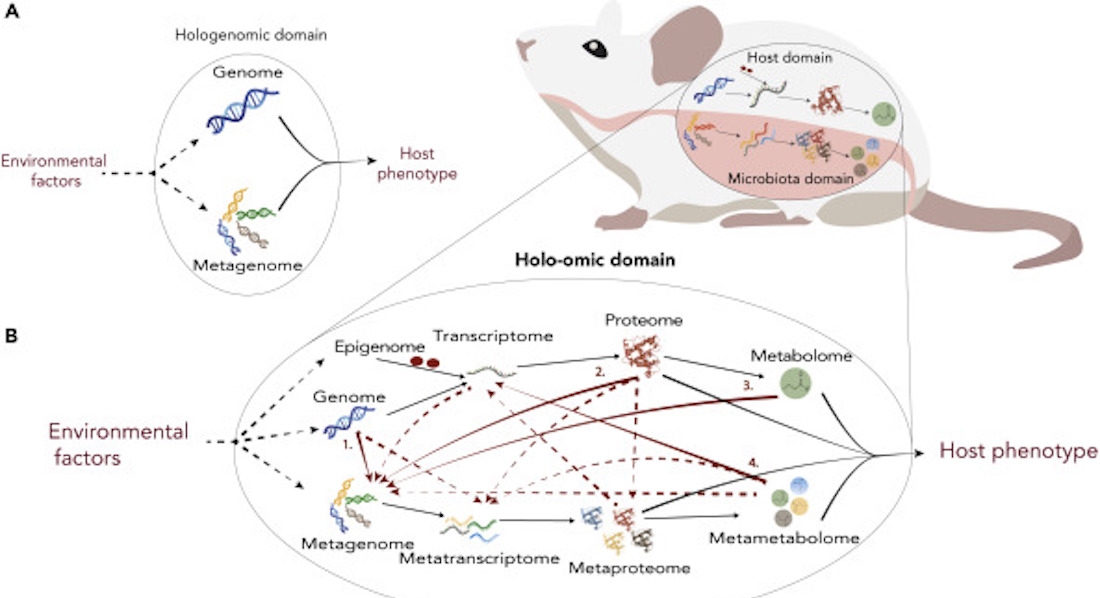
Guideline on how to apply the holo-omic approach
The authors guide researchers through applying the holo-omic approach to new research fields by describing how the holo-omic toolbox can be used to study host-microbiota interactions at various levels of complexity providing three main questions that researchers need to consider to maximise the outcome of a holo-omic study.
The three main guiding questions for researchers to consider are: 1) are host-microbiota interactions relevant in the study system? 2) is it meaningful to implement a holo-omic approach? 3) which omic levels are relevant and how to maximise the amount of useful data derived from them? The questions and guidelines are expanded and explained further in the paper.
Improving health for both chicken and humans
Furthermore, the perspective paper provides examples within each field of research in order to showcase the potential provided by the new holo-omic approach to host-microbiota interactions within each of the fields of biomedicine, biotechnology, agri- and aquacultural sciences and nature conservation.
One such example is from agricultural sciences, where the need for more sustainable and healthy poultry production can be achieved by taking into account the entire biochemical landscape to inform better designed feed additive strategies and in turn decrease production inefficiency driven by gut dysbiosis.
Incorporating the holo-omics approach to biomedical research offers another exciting avenue towards better treatments of many modern human diseases including holo-omics based personalised medicine which would not only recognise the genetic and exposomic features of patients, but also the associated microbiota.
Holo-omics: a solution for the future
Finally, the authors end the perspective by identifying the limiting factors that currently prevent the widespread implementation of the holo-omic approach and discuss possible solutions to overcome them. Although at present the widespread application of holo-omics remains elusive due to technical, financial and biological limitations, there are future solutions that will overcome these. The increase of publications, tools and holo-omic data sets will only aid this and it will not be long before holo-omics is applied to an even broader range of research questions.
Read more in the original open access paper: https://doi.org/10.1016/j.isci.2020.101414
Contact:
